Fluorescence in Situ Hybridization (FISH)
Fluorescent in situ hybridization (FISH) is a powerful technique for detecting RNA or DNA sequences in cells, tissues, and tumors. FISH provides a unique link among the studies of cell biology, cytogenetics, and molecular genetics.
Fluorescent in situ hybridization is a technique in which single-stranded nucleic acids (usually DNA, but RNA may also be used) are permitted to interact so that complexes, or hybrids, are formed by molecules with sufficiently similar, complementary sequences. Through nucleic acid hybridization, the degree of sequence identity can be determined, and specific sequences can be detected and located on a given chromosome.
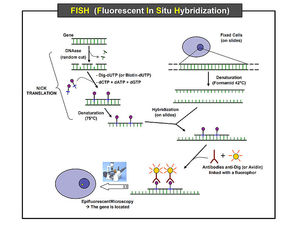
The method comprises of three basic steps: fixation of a specimen on a microscope slide, hybridization of labeled probe to homologous fragments of genomic DNA, and enzymatic detection of the tagged target hybrids. While probe sequences were initially detected with isotopic reagents, nonisotopic hybridization has become increasingly popular, with fluorescent hybridization now a common choice. Protocols involving nonisotopic probes are considerably faster, with greater signal resolution, and provide options to visualize different targets simultaneously by combining various detection methods.
The detection of sequences on the target chromosomes is performed indirectly, commonly with biotinylated or digoxigenin-labeled probes detected via a fluorochrome-conjugated detection reagent, such as an antibody conjugated with fluorescein. As a result, the direct visualization of the relative position of the probes is possible. Increasingly, nucleic acid probes labeled directly with fluorochromes are used for the detection of large target sequences. This method takes less time and results in lower background; however, lower signal intensity is generated. Higher sensitivity can be obtained by building layers of detection reagents, resulting in amplification of the signal. Using such means, it is possible to detect single-copy sequences on chromosome with probes shorter than 0.8 kb.
Probes can vary in length from a few base pairs for synthetic oligonucleotides to larger than one Mbp. Probes of different types can be used to detect distinct DNA types. PCR-amplified repeated DNA sequences, oligonucleotides specific for repeat elements, or cloned repeat elements can be used to detect clusters of repetitive DNA in heterochromatin blocks or centromeric regions of individual chromosomes. These are useful in determining aberrations in the number of chromosomes present in a cell. In contrast, for detecting single locus targets, cDNAs or pieces of cloned genomic DNA, from 100 bp to 1 Mbp in size, can be used.
To detect specific chromosomes or chromosomal regions, chromosome-specific DNA libraries can be used as probes to delineate individual chromosomes from the full chromosomal complement. Specific probes have been commercially available for each of the human chromosomes since 1991.
Any given tissue or cell source, such as sections of frozen tumors, imprinted cells, cultured cells, or embedded sections, may be hybridized. The DNA probes are hybridized to chromosomes from dividing (metaphase) or non-dividing (interphase) cells.
The observation of the hybridized sequences is done using epifluorescence microscopy. White light from a source lamp is filtered so that only the relevant wavelengths for excitation of the fluorescent molecules reach the sample. The light emitted by fluorochromes is generally of larger wavelengths, which allows the distinction between excitation and emission light by means of a second optical filter. Therefore, it is possible to see bright colored signals on a dark background. It is also possible to distinguish between several excitation and emission bands, thus between several fluorochromes, which allows the observation of many different probes on the same target.
FISH has a large number of applications in molecular biology and medical science, including gene mapping, diagnosis of chromosomal abnormalities, and studies of cellular structure and function. Chromosomes in three-dimensionally preserved nuclei can be "painted" using FISH. In clinical research, FISH can be used for prenatal diagnosis of inherited chromosomal aberrations, postnatal diagnosis of carriers of genetic disease, diagnosis of infectious disease, viral and bacterial disease, tumor cytogenetic diagnosis, and detection of aberrant gene expression. In laboratory research, FISH can be used for mapping chromosomal genes, to study the evolution of genomes (Zoo FISH), analyzing nuclear organization, visualization of chromosomal territories and chromatin in interphase cells, to analyze dynamic nuclear processes, somatic hybrid cells, replication, chromosome sorting, and to study tumor biology. It can also be used in developmental biology to study the temporal expression of genes during differentiation and development. Recently, high resolution FISH has become a popular method for ordering genes or DNA markers within chromosomal regions of interest.
See also Chromosome mapping; Genetic engineering; Genetic testing.
Resources
Books
Spector, D.L., R.D. Goldman, and L.A. Leinwand. Cells: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press, 1998.
Periodicals
Nath, J., et al. "A Review of Fluorescence in situ Hybridization (FISH): Current Status and Future Prospects." Biotech Histochem. 75 (March 2000): 54-78.
Nicole D. Le Brasseur
Additional topics
Science EncyclopediaScience & Philosophy: Ferroelectric materials to Form and matter